Doctoral studies (2011-2016)
I completed my PhD training (2016) in the department of Developmental and Cell Biology and the Center for Complex Biological Systems at the University of California Irvine with Dr. Ali Mortazavi. My thesis was focused on mapping the dynamic gene regulatory networks during human myeloid differentiation using ATAC-seq and RNA-seq data (Ramirez et al, 2017 Cell Systems). My training included the integration of molecular biology, biochemistry, next-generation sequencing (genomic technologies) and computational approaches.
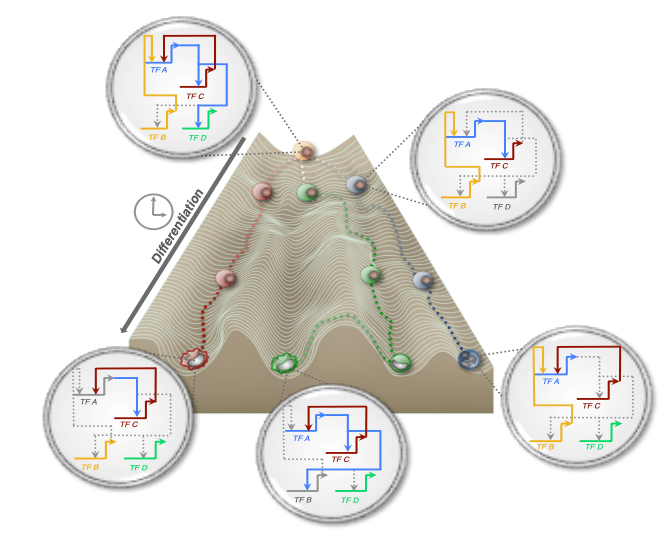
Macrophage/Monocyte dynamic gene regulatory networks
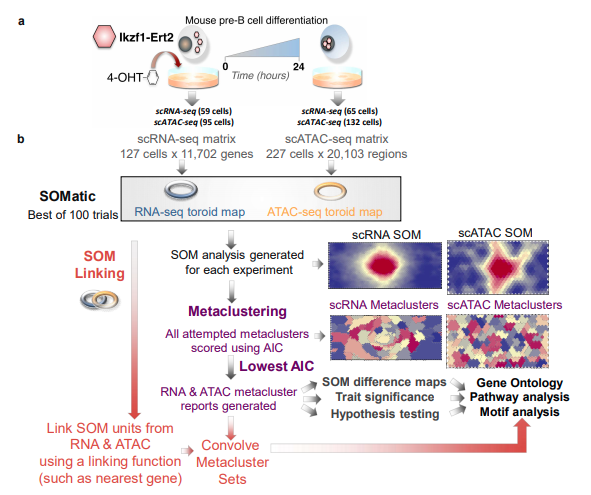
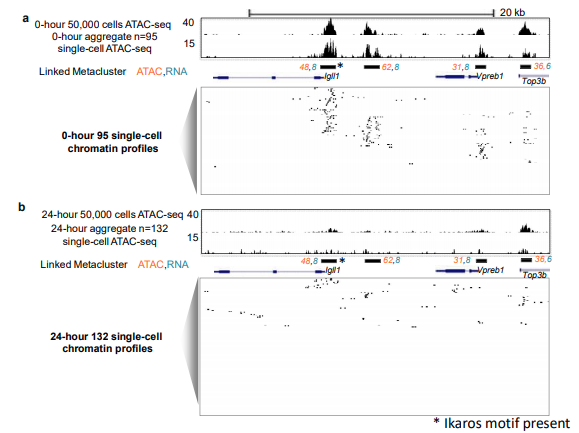
I participated actively in the STATegra consortium, which mapped molecular dynamics during pre-B cell differentiation using multi-omic data integration (STATegra et al, Scientific Data 2019). My role in STATegra was primarily geared towards data generation and analysis of bulk DNase/ATAC-seq, single-cell RNA-seq, and single-cell ATAC-seq across the pre-B model system. Along with my colleague Camden Jansen, we developed a novel approach using self-organizing maps (SOM) to link scATAC-seq and scRNA-seq data that overcomes these challenges and can generate draft regulatory networks (Jansen, Ramirez et al 2019).
Building GRNs using Self-Organizing Maps (SOM)
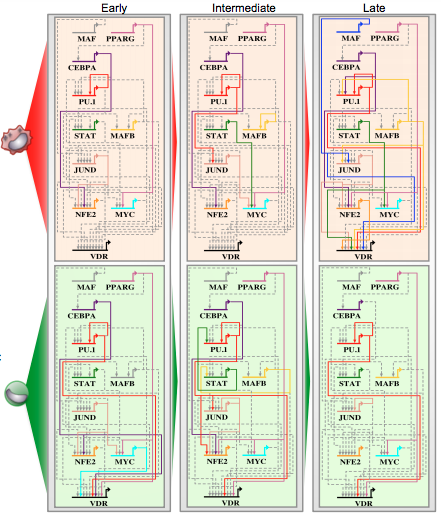
During my PhD I was an active member of the ENCODE consortium. Aside from my own thesis work, I was fortunate enough to participate in several research collaborations during my PhD. This included studies on myofibroblast regeneration (Plikus et al 2017 Science. Maksim Plikus lab, UCI), derivation of microglia from human iPSC (Abud et al, Neuron 2017), and transcriptional regulation across several cellular systems (Peter Donovan lab, UCI; Esteban Ballestar lab (Rodriguez-Ubreva et al 2017 Cell Reports).
Postdoctoral fellowship studies (2017-2021)
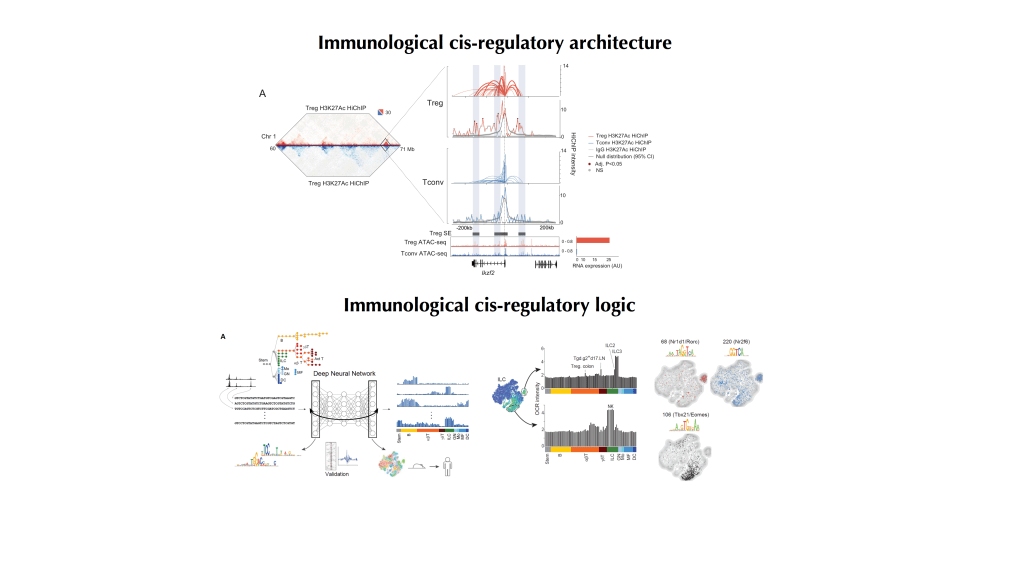
As an NIH funded postdoctoral fellow in the laboratory of Dr. Christophe Benoist and Dr. Diane Mathis at Harvard Medical School in the Department of Immunology and member of the ImmGen consortium I participated in the analysis of the Cis-regulatory Atlas of the mouse immune system (Yoshida et al 2019 Cell) and the focused analysis of the immune cis-regulatory syntax using deep learning approaches (Maslova, Ramirez et al 2020 PNAS). In collaboration with Hur lab, I worked on the epigenomic analysis of FoxP3 dimerization (Leng et al Immunity 2022) and its’ role in DNA bridging via multimerization (Zhang et al Nature 2023). My primary focus was to study the the 3D genome architecture of regulatory T cells and the role of FoxP3-mediated looping as they relate to the regulation of Treg gene programs (Ramirez et al Science Immunology 2022).
Computational software and applications

An R based visualizer and network analysis of chromatin conformation data
Online web app: http://shiny.immgen.org/DNARchitect/
Github: https://github.com/alosdiallo/DNA_Rchitect
RNR and AD conceived the idea for DNA Rchitect.
DNA Rchitect was built and developed with Alos Diallo, Karni Bedirian and Simon Gray
2021-2025
- Associate Director of Data Sciences, nChroma Bio (formerly Chroma Medicine). Boston MA. (current)
- Sr. Scientist Computational Biology in Data Sciences at Chroma Medicine. Boston MA. (April 2022-December 2023)
- Scientist II Computational Biology in Data Sciences at Chroma Medicine. Boston MA. (June 2021-April 2022)
2026-Current
- Chief Scientific Officer of the MED13L Foundation.
- Founder of RNR Consulting LLC.

- Scientific Advisory Board Member to NeuCyte Inc.
- Medical Advisory Board Member to Koolen-de Vries Foundation.
Additional Scientific interests
Apart from work in the innate and adaptive immune systems, I am also interested in studying various common and rare disease pathologies in the brain. This extends to understanding how different gene regulatory programs of the same cell type become active in different disease contexts. Moreover development of genomic/imaging technologies and computational software for interrogating gene regulation and various chromatin features are areas of keen interest.
If you would like to contact me, please email me at ricardo.ramirez.hms@gmail.com.
Non-science related interests
I play electric/acoustic guitar and listen to music ranging from french house, jazz to electronic. I enjoy keeping fit, hiking, martial arts, and being adventurous. I enjoy saltwater and freshwater fishing, and spending time outdoors. I relish in exploring and spending quality time with loved ones and close friends.
Complete list of publications: Google Citations
Publications
* Denotes equal contributions
2026
26) Regulatory T cells safeguard liver health during metabolic-associated steatohepatitis. Bola Hanna, {…}, Ricardo N. Ramirez, {…}, Diane Mathis. (PNAS 2026)
2025
25) Uncovering the regulatory landscape of early human early B-cell lymphopoiesis and its implications in the pathogenesis of B-ALL. Núria Planell*, Xabier Martínez, Daniel Mouzo, David Lara-Astiaso, Amaia Vilas-Zornoza, Patxi San Martín-Uriz, Diego Alignani, Bruno Paiva, Alberto Maillo, Aleksandra Kurowska, Arantxa Urdangarin, Vincenzo Lagani, Asier Ortega, Mikel Hernaez, Narsis Kiani, Jose Martin-Subero, Ricardo N. Ramirez*, Jesper Tegner*, Felipe Prosper*, David Gomez-Cabrero*. (Science Advances 2025).
24) Iterative transcription factor screening enables rapid generation of microglia-like cells from human iPSC. Songlei Liu, {…}*, Ricardo N. Ramirez, {…}, Soumya Raychaudhuri#, George M. Church#. (Nature Communications 2025).
23) A potent epigenetic editor targeting human PCSK9 for durable reduction of low-density lipoprotein cholesterol levels. Frederic Tremblay, {…} , Ricardo N. Ramirez, {…}, Aron B. Jaffe. (Nature Medicine 2025)
2023
22) FOXP3 recognizes microsatellites and bridges DNA through multimerization. Wenxiang Zhang, Fangwei Leng, Xi Wang, Ricardo N. Ramirez, Jinseok Park, Christophe Benoist, Sun Hur. (Nature 2023).
21) Mutations from IPEX patients ported to mice reveal different patterns of FoxP3 and Treg dysfunction. Juliette Leon, Kaitavjeet Chowdhary, Wenxiang Zhang, Ricardo N. Ramirez, Isabelle Andre, Sun Hur, Diane Mathis, Christophe Benoist. (Cell Reports 2023).
20) The gut microbiota promotes distal tissue regeneration via RORγ+ regulatory T cell emissaries. Bola Hanna, Gang Wang, Silvia Galvan-Pena, Alexander O. Mann, Ricardo N. Ramirez, Andres Munoz-Rojas, Kathleen Smith, Min Wan, Christophe Benoist, Diane Mathis. (Immunity 2023).
2022
19) The transcription factor FoxP3 can fold into two distinct dimerization states with divergent functional implications for Treg homeostasis. Fangwei Leng, Wenxiang Zhang, Ricardo N. Ramirez, Juliette Leon, Yi Zhong, Joris van der Veeken, Alexander Rudensky, Christophe Benoist, Sun Hur. (Immunity 2022).
18) FoxP3 associates with enhancer-promoter loops to regulate Treg-specific gene expression. Ricardo N. Ramirez, Kaitavjeet Chowdhary, Juliette Leon, Diane Mathis, Christophe Benoist. (Science Immunology 2022).
17) Maternal gut bacteria drives intestinal inflammation in offspring with neurodevelopmental disorders by altering the chromatin landscape of CD4+ T cells. Eunha Kim, Donggi Paik, Ricardo N. Ramirez, Delaney G. Biggs, Youngjun Park, Ho-Keun Kwon, Gloria B. Choi, Jun R. Huh. (Immunity 2022).
2021
16) Aire regulates chromatin looping by evicting CTCF from domain boundaries and favoring accumulation of cohesin on super-enhancers. Kushagra Bansal, Daniel A. Michelson, Ricardo N. Ramirez, Aaron D. Viny, Ross L. Levine, Christophe Benoist, Diane Mathis. (PNAS 2021).
15) Interferon-α-producing plasmacytoid dendritic cells drive the loss of adipose tissue regulatory T cells during obesity. Chaoran Li, Gang Wang, Pulavendran Sivasami, Ricardo N. Ramirez, Yanbo Zhang, Christophe Benoist, and Diane Mathis. (Cell Metabolism 2021).
2020
14) Deep learning of immune cell differentiation. Alexandra Maslova*, Ricardo N. Ramirez*, Ke Ma, Hugo Schmutz, Chendi Wang, Curtis Fox, Bernard Ng, Christophe Benoist#, Sara Mostafavi#, and the Immunological Genome Project Consortium. #Joint supervision. (PNAS 2020).
13) Perspectives on ENCODE. The ENCODE Project Consortium (Nature 2020).
12) Expanded encyclopaedias of DNA elements in the human and mouse genomes. The ENCODE Project Consortium (Nature 2020).
11) Slug regulates the Dll4-Notch,VEGFR2 axis to control endothelial cell activation and angiogenesis. Nan Hultgren, Jennifer Fang, Mary Ziegler, Ricardo N. Ramirez, Duc Phan, Michaela Hatch, Katrina Welch-Reardon, Antonio Paniagua, Lin Kim, Nathan Shon, Ali Mortazavi, David Williams, Christopher Hughes. (Nature Communications 2020).
2019
10) DNA Rchitect: An R based visualizer for network analysis of chromatin interaction data. Ricardo N. Ramirez*, Karni Bedirian*, Simon Gray, Alos Diallo. (Bioinformatics 2019).
9) The cis-regulatory atlas of the mouse immune system. Hideyuki Yoshida, Caleb A. Lareau, Ricardo N. Ramirez, Samuel A. Rose, Barbara Maier, Aleksandra Wroblewska. Fiona Desland, Aleksey Chudnovskiy, Arthur Mortha, Claudia Dominguez, Julie Tellier, Edy Kim, Dan Dwyer, Susan Shinton, Tsukasa Nabekura, YiLin Qi, Bingfei Yu, Michelle Robinette, Kiwook Kim, Amy Wagers, Andrew Rhoads, Stephen Nutt, Brian Brown, Sara Mostafavi, Jason Buenrostro, Christophe Benoist, and the Immunological Genome Project Consortium. (Cell 2019).
8) Building gene regulatory networks from single-cell ATAC-seq and RNA-seq using Linked Self-Organizing Maps. C*, *, ,
7) Feedforward regulation of Myc coordinates lineage-specific with housekeeping gene expression during B cell progenitor cell differentiation. Isabel Ferreirós Vidal, Thomas Carroll, Tianyi Zhang, Vincenzo Lagani, Ricardo N. Ramirez, Elizabeth Ing-Simmons, Alicia Garcia, Lee Cooper, Ziwei Liang, Georgios Papoutsoglou, Gopuraja Dharmalingam, Ya Guo, Sonia Tarazona, Sunjay J. Fernandes, Peri Noori, Gilad Silberberg, Amanda G. Fisher, Ioannis Tsamardinos, Ali Mortazavi, Boris Lenhard, Ana Conesa , Jesper Tegner, Matthias Merkenschlager, David Gomez-Cabrero (PLOS Biology 2019).
6) STATegra: a comprehensive multi-omics dataset of B-cell differentiation in mouse.
David Gomez-Cabrero*, Sonia Tarazona*, Isabel Ferreirós-Vidal*, Ricardo N. Ramirez*, Carlos Company*, Andreas Schmidt*, Theo Reijmers*, Veronica von Saint Paul*, Franscesco Marabita, Javier Rodríguez-Ubreva, Antonio Garcia-Gomez, Thomas Carroll, Lee Cooper, Ziwei Liang, Gopuraja Dharmalingam, Leandro Balzano, Vicenzo Lagani, Ioannis Tsamardinos, Michael Lappe, Dieter Maier, Johan A. Westerhuis, Thomas Hankemeier, Axel Imhof, Esteban Ballestar, Ali Mortazavi, Matthias Merkenschlager, Jesper Tènger, Ana Conesa. (Scientific Data 2019).
2018
5) TCF7L1 suppresses primitive streak gene expression to support human embryonic stem cell pluripotency. Robert A. Sierra, Nathan P. Hoverter, Ricardo N. Ramirez, Linh M. Vuong, Ali Mortazavi, Bradley J. Merrill, Marian L. Waterman, Peter J. Donovan (Development 2018).
2017
4) Dynamic gene regulatory networks of human myeloid differentiation. Ricardo N. Ramirez,{…}, Ali Mortazavi (Cell Systems 2017).
3) iPSC-derived human microglia-like cells to study neurological disease. E.M. Abud1,2,3, R.N. Ramirez4,5, E.S. Martinez1,2,3, L.M. Healy7, C.H.H. Nguyen1,2,3, V.M. Scarfone2, S.E. Marsh1,2,3, A. Madany8, C. Fimbres2,3, C. Caraway2,3, M.D. Torres1,2,3, A. Park2,3, K. Rakez6, A., K.H. Gylys7, A. Mortazavi4,5, J.P. Antel7, M.J. Carson2, W.W. Poon2,3, M. Blurton-Jones. (Neuron 2017).
2) Regeneration of fat cells from myofibroblasts during wound healing. Maksim V. Plikus1,2, Christian Fernando Guerrero-Juarez2, Mayumi Ito3, Yun Rose Li4, Priya H. Dedhia5, Olga Shestova5, Denise L. Gay1,6, Raul Ramos2, Tsai-Ching Hsi2, Ji Won Oh2, Xiaojie Wang2, Ricardo N. Ramirez, Rabi Murad, Amanda Ramirez2, Sara E. Konopelski2, Arijh Elzein2, Anne Wang1, Rarinthip June Supapannachart1, Zaixin Yang1, Ying Zheng1, Arben Nace1, Amy Guo1, Elsa Treffeisen1, Thomas Andl7, Stefan Offermanns8, Daniel Metzger9, Pierre Chambon9, Ali Mortazavi, Rana K. Gupta10, Bruce A. Hamilton11, Sarah E. Millar1, Patrick Seale5, 12, Warren S. Pear5, Mitchell A. Lazar4, 13, George Cotsarelis (Science, 2017).
1) Prostaglandin E2 Leads to the Acquisition of DNTM3A-Dependent Tolerogenic Functions in Human Myeloid-Derived Suppressor Cells. Javier Rodríguez-Ubreva,Francesc Català-Moll, Nataša Obermajer, Damiana Álvarez-Errico, Ricardo N. Ramirez, Carlos Company, Roser Vento-Tormo, Gema Moreno-Bueno, Robert P. Edwards, Ali Mortazavi, Pawel Kalinski, Esteban Ballestar. (Cell Reports, 2017).